Append a new dataset¶
We have one dataset in storage and are about to receive a new dataset.
In this notebook, we’ll see how to manage the situation.
import lamindb as ln
import bionty as bt
import readfcs
bt.settings.organism = "human"
ln.track("SmQmhrhigFPL0000")
→ connected lamindb: testuser1/test-facs
→ created Transform('SmQmhrhigFPL0000'), started new Run('OKzq4Ixx...') at 2025-07-14 06:43:53 UTC
→ notebook imports: bionty==1.6.0 lamindb==1.8.0 pytometry==0.1.6 readfcs==2.0.1 scanpy==1.11.3
Ingest a new artifact¶
Access  ¶
¶
Let us validate and register another .fcs file from Oetjen18:
filepath = readfcs.datasets.Oetjen18_t1()
adata = readfcs.read(filepath)
adata
Transform: normalize  ¶
¶
import pytometry as pm
pm.pp.split_signal(adata, var_key="channel")
pm.pp.compensate(adata)
pm.tl.normalize_biExp(adata)
adata = adata[ # subset to rows that do not have nan values
adata.to_df().isna().sum(axis=1) == 0
]
adata.to_df().describe()
Validate cell markers  ¶
¶
Let’s see how many markers validate:
validated = bt.CellMarker.validate(adata.var.index)
Let’s standardize and re-validate:
adata.var.index = bt.CellMarker.standardize(adata.var.index)
validated = bt.CellMarker.validate(adata.var.index)
Next, register non-validated markers from Bionty:
records = bt.CellMarker.from_values(adata.var.index[~validated])
ln.save(records)
Manually create 1 marker:
bt.CellMarker(name="CD14/19").save()
Move metadata to obs:
validated = bt.CellMarker.validate(adata.var.index)
adata.obs = adata[:, ~validated].to_df()
adata = adata[:, validated].copy()
Now all markers pass validation:
validated = bt.CellMarker.validate(adata.var.index)
assert all(validated)
Register  ¶
¶
curate = ln.Curator.from_anndata(adata, var_index=bt.CellMarker.name, categoricals={})
curate.validate()
artifact = curate.save_artifact(description="Oetjen18_t1")
Annotate with more labels:
efs = bt.ExperimentalFactor.lookup()
organism = bt.Organism.lookup()
artifact.labels.add(efs.fluorescence_activated_cell_sorting)
artifact.labels.add(organism.human)
artifact.describe()
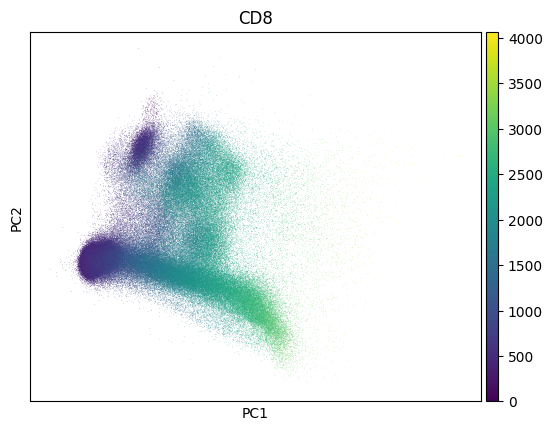
Inspect a PCA fo QC - this collection looks much like noise:
import scanpy as sc
markers = bt.CellMarker.lookup()
sc.pp.pca(adata)
sc.pl.pca(adata, color=markers.cd8.name)
Create a new version of the collection by appending a artifact¶
Query the old version:
collection_v1 = ln.Collection.get(key="My versioned cytometry collection")
collection_v2 = ln.Collection(
[artifact, collection_v1.ordered_artifacts[0]],
revises=collection_v1,
version="2",
)
collection_v2.describe()
collection_v2.save()